-
Notifications
You must be signed in to change notification settings - Fork 2
Graph Algorithms
Simple Graph : Graph with no self loops or multi-edges.
Incident Edges
: Edges coming off a node. Degree(v) is the number of Incident Edges.
Number of Vertices:
Min num edges in an Unconnected Graph = 0
Min num edges in a Minimally Connnected Graph =
Max num edges in Simple Graph =
Relationship between degree of graph and edges:
Graph Implementations
-
Edge List
An array of pairs of vertices (Edges). If edges have weights, add either a third element to the array or more information to the object.
The total space for an edge list is Θ(E).
To find if the graph contains a particular edge, we have to search through the entire edge list, so O(|E|).
Example :
[[0,1], [0,6], [0,8], [1,4], [1,6], [1,9], [2,4], [2,6], [3,4], [3,5]]Operation Run Time insertVertex(K key) $O(1)$ removeVertex(K key) $O(m)$ areAdjacent(Vertex v1, Vertex v2) $O(m)$ incidentEdges(Vertex v) $O(m)$ -
Adjacency Matrix
v = Number of vertices (
|V|).The matrix is
v x v. Number of edges is irrelevant.An adjacency matrix can determine whether two vertices are adjacent in
$O(1)$ time by looking at the corresponding entry.matrix[i][j]is 1 if the edge(i,j)exists in the graph, otherwise it's 0.The diagonal should be all 0s if the graph is Simple (no self edges).
For an undirected graph, adjacency matrix is symmetric. For a directed graph, adjacency matrix is not necessarily symmetric.
It takes Θ(V2) space, even if the graph is sparse.
Example:
[0, 1, 0, 0, 0, 0] [1, 0, 0, 0, 1, 0] [0, 0, 0, 0, 1, 0] [0, 0, 0, 0, 1, 1] [0, 1, 1, 1, 0, 1] [0, 0, 0, 1, 1, 0]Operation Run Time insertVertex(K key) Amortized $O(n)*$ (?)removeVertex(K key) $O(n)$ areAdjacent(Vertex v1, Vertex v2) O(1) incidentEdges(Vertex v) $O(n)$ -
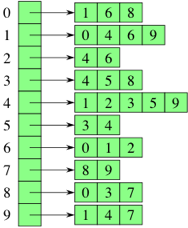
Adjacency List
Combines adjacency matrices with edge lists.
For each vertex, store an array of adjacenct vertices. Max size of array is
$n - 1$ .$2m$ list nodes. (?)Operation Run Time insertVertex(K key) O(1) removeVertex(K key) O(deg(v)) areAdjacent(Vertex v1, Vertex v2) O( min(deg(v1), deg(v2)) ) incidentEdges(Vertex v) O(deg(v))
Graph BFS/DFS
Both BFS and DFS have optimal run times of
For a DFS, just switch the Queue to a Stack.
BFS -- "CROSS" DFS -- "BACK"
void initBFS(Graoh g_) {
// Set all vertex labels to "UNEXPLORED"
for (auto v : g_.getVertices()) {
g_.setVertexLabel(v, "UNEXPLORED");
}
// Set all edge labels to "UNEXPLORED"
for (auto e : g_.getEdges()) {
g_.setEdgeLabel(e, "UNEXPLORED");
}
// Runs BFS on all vertices
for (auto v : g_.getVertices()) {
if (g_.getLabel(v) == "UNEXPLORED") {
BFS(g_, v);
}
}
}
void BFS(Graph g_, Vertex v) {
Queue<Vertex> q;
g_.setLabel(v, "EXPLORED");
q.push(v);
while (! q.empty()) {
Vertex next = q.front();
q.pop();
for (auto adjV : g_.getAdjacentVertices(next)) {
if (g_.getLabel(adjV) == "UNEXPLORED") {
g_.setLabel(next, adjV, "DISCOVERY");
q.push(adjV);
} else g_.setLabel(next, adjV, "BACK");
}
}
}Spanning Tree : Every possible vertex in G is connected with minimum number of edges. Connected and acyclic.
Minimum Spanning Tree
: Spanning tree with the minimal total edge weights. Has
Partition Property : If there's an edge of minimum weight across the partition, it's a part of the MST.
To run MST algorithm, underlying graph must be connected. So,
Fibionacci Heap
: Decreases value of key while maintaining heap property in Amortized
Negative weight cycles break everything.
Both Kruskal and Prim's run in
- Kruskal's Algorithm (MST) -- Next lowest weight edge until MST formed
- Choose the shortest edge
- Choose the next lowest weighted edge on the graph, anywhere, that wouldn't create a cycle. Do this over and over until every vertex is in the tree.

| Priority Queue Implementation | Total Run Time |
|---|---|
| Heap | |
| Sorted Array |
void KruskalMST(Graph& graph) {
// Get a list of all edges in the graph and sort them by increasing weight.
queue<Edge> queue;
std::vector<Edge> edges = graph.getEdges();
std::sort(edges.begin(), edges.end());
for (auto edgesIter = edges.begin(); edgesIter != edges.end(); edgesIter++) {
queue.push(*edgesIter);
}
// Create a disjoint sets structure where each vertex is represented by a set
DisjointSets forest;
std::vector<Vertex> vertices = graph.getVertices();
int numVertices = vertices.size();
forest.addelements(vertices.size());
/**
* Traverse the list from the start (i.e., from lightest weight to heaviest).
* Inspect the current edge. If that edge connects two vertices from different sets,
union their respective sets and mark the edge as part of the MST.
Otherwise there would be a cycle, so do nothing.
* Repeat this until n−1 edges have been added, where n is the number of vertices in the graph.
*/
int numEdgesAdded = 0;
while (numEdgesAdded < numVertices - 1) {
Edge nextEdge = queue.front();
queue.pop();
Vertex source = nextEdge.source;
Vertex dest = nextEdge.dest;
if (forest.find(source) != forest.find(dest)) {
graph.setEdgeLabel(source, dest, "MST");
forest.setunion(source, dest);
numEdgesAdded++;
}
}
}- Prim's Algorithm (MST) -- Partition Algorithm.
- Choose start vertex.
- Add the next lowest weighted incident edge that wouldn't create a cycle and is not already visited. Do this over and over until you have an MST.
 https://www.youtube.com/watch?v=cplfcGZmX7I
https://www.youtube.com/watch?v=cplfcGZmX7I
void Prim(Graph& g, Vertex start) {
vector<int> dist;
vector<Vertex> predecessor;
//Set all initial distances to max, predecessors to NULL.
for (int i = 0; i < g.getVertices().size(); i++) {
dist[i] = +inf ***(use 99999);
predecessor[i] = NULL
}
dist[start] = 0;
Queue<Vertex> queue;
}
| Implementation | Adj. Matrix | Adj. List |
|---|---|---|
| Heap | ||
| Unsorted Array |
Shortest Path Algorithms
-
Dijkstra's Algorithm -- Closely related to Prim's
- Can look up distance to any node in
$O(1)$ time - Every edge needs a non-negative weight.
- Next vertex weight is the previous vertex weight plus the edge weight to get there.
- https://www.youtube.com/watch?v=_lHSawdgXpI
-
$O(m + nlog(n))$ when using Fibionnaci Trees (My notes say this is the same as Prim's?)
- Can look up distance to any node in
-
Floyd-Warshall
- Similar to Dijkstra, but can be used with negative edge weights.
- Time Complexity of
$O(n^{3})$